Introduction to Single-Cell RNA Sequencing
RNA sequencing (RNA-seq) is a genomic technique for detecting and quantifying messenger RNA molecules in biological samples, which is useful for studying cellular responses. In recent years, RNA-seq has fueled a lot of medical discoveries and innovations. The approach is frequently applied to samples containing thousands to millions of cells for practical reasons. However, direct evaluation of biology's fundamental unit, the cell, has been hampered as a result of this. Many more studies have been released since the first single-cell RNA-sequencing (scRNA-seq) study in 2009, mostly by specialist laboratories with special abilities in wet-lab single-cell genomics, bioinformatics, and computation. With the growing commercial accessibility of scRNA-seq platforms and the rapid maturation of bioinformatics methods, any biomedical researcher or clinician can now use scRNA-seq to make interesting discoveries.
Advantages and Applications of Single-Cell RNA Sequencing
Individual cell transcriptomes can be compared using scRNA-seq. As a result, one of the most common applications of scRNA-seq has been to assess transcriptional similarities and differences within a population of cells, with early reports exposing previously unappreciated levels of heterogeneity in embryonic and immune cells, for example. As a result, heterogeneity analysis remains a primary motivation for pursuing scRNA-sequencing studies.
Similarly, transcriptional distinctions between individual cells have been utilized to define rare cell populations that would otherwise go undetected in pooled cell analyses, such as malignant tumor cells within a tumor mass or hyper-responsive immune cells within an otherwise homogeneous group. Individual T lymphocytes expressing highly diverse T-cell receptors, neurons in the brain, or cells in an early-stage embryo are all good candidates for scRNA-seq analysis. In situations such as embryonal development, cancer, myoblast and lung epithelium differentiation, and lymphocyte fate diversification, scRNA-seq is progressively being used to detect lineage and developmental relationships between heterogeneous, yet related, cellular states.
Please read our article Single-Cell RNA Sequencing in Drug Discovery and Development for more opportunities.
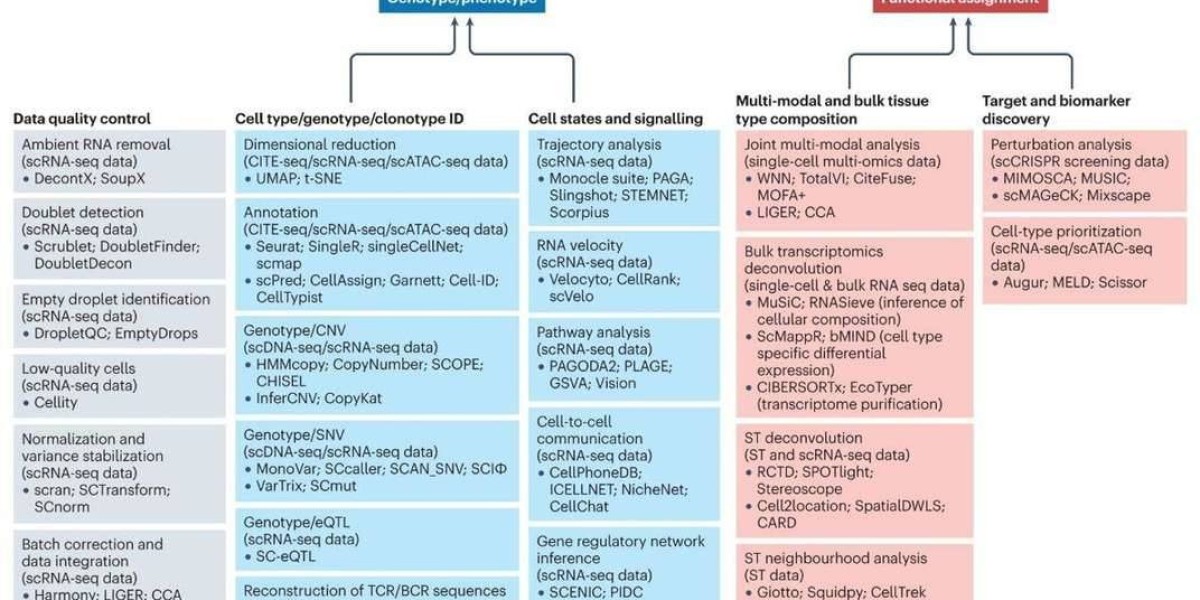
Methods and Technologies for Single-Cell RNA Sequencing
Even though many scRNA-seq studies have reported bespoke techniques, such as new developments in wet-lab, bioinformatics, or computational tools, the majority have followed a standard methodological pipeline. The efficient isolation of viable, single cells from the tissue of interest was the first and most essential step in conducting scRNA-seq. However, emerging technologies such as isolation of single nuclei for RNA-seq and 'split-pooling' scRNA-seq strategies based on combinatorial indexing of single cells, which are based on combinatorial indexing of single cells, offer some advantages over isolation of single intact cells, such as enabling for easier analyses of fixed samples and ignoring the need for expensive hardware.
Individual cells are then lysed in order to acquire as many RNA molecules as possible. Poly[T]-primers are frequently used to particularly analyze polyadenylated mRNA molecules while avoiding capturing ribosomal RNAs. Non-polyadenylated mRNA analysis is usually more difficult and necessitates the use of specialized protocols. A reverse transcriptase converts poly[T]-primed mRNA to complementary DNA (cDNA). Other nucleotide sequences, such as adaptor sequences for detection on NGS platforms, unique molecular identifiers to define unequivocally a single mRNA molecule, and sequences to maintain information on the cellular origin, will be added to the reverse-transcription primers based on the scRNA-seq protocol. The cDNA is then amplified by PCR or, in some cases, in vitro transcription accompanied by another round of reverse transcription—some protocols opt for nucleotide barcode-tagging at this stage to maintain information on cellular origin. The amplified and tagged cDNA from each cell is then pooled and sequenced by NGS, using library preparation methods, sequencing platforms, and genomic-alignment tools that are related to those used with bulk specimens.
References:
- Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nature Reviews Immunology. 2018 Jan;18(1).
- Kolodziejczyk AA, Kim JK, Svensson V, et al. The technology and biology of single-cell RNA sequencing. Molecular cell. 2015 May 21;58(4).
- Wu AR, Neff NF, Kalisky T, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nature methods. 2014 Jan;11(1).